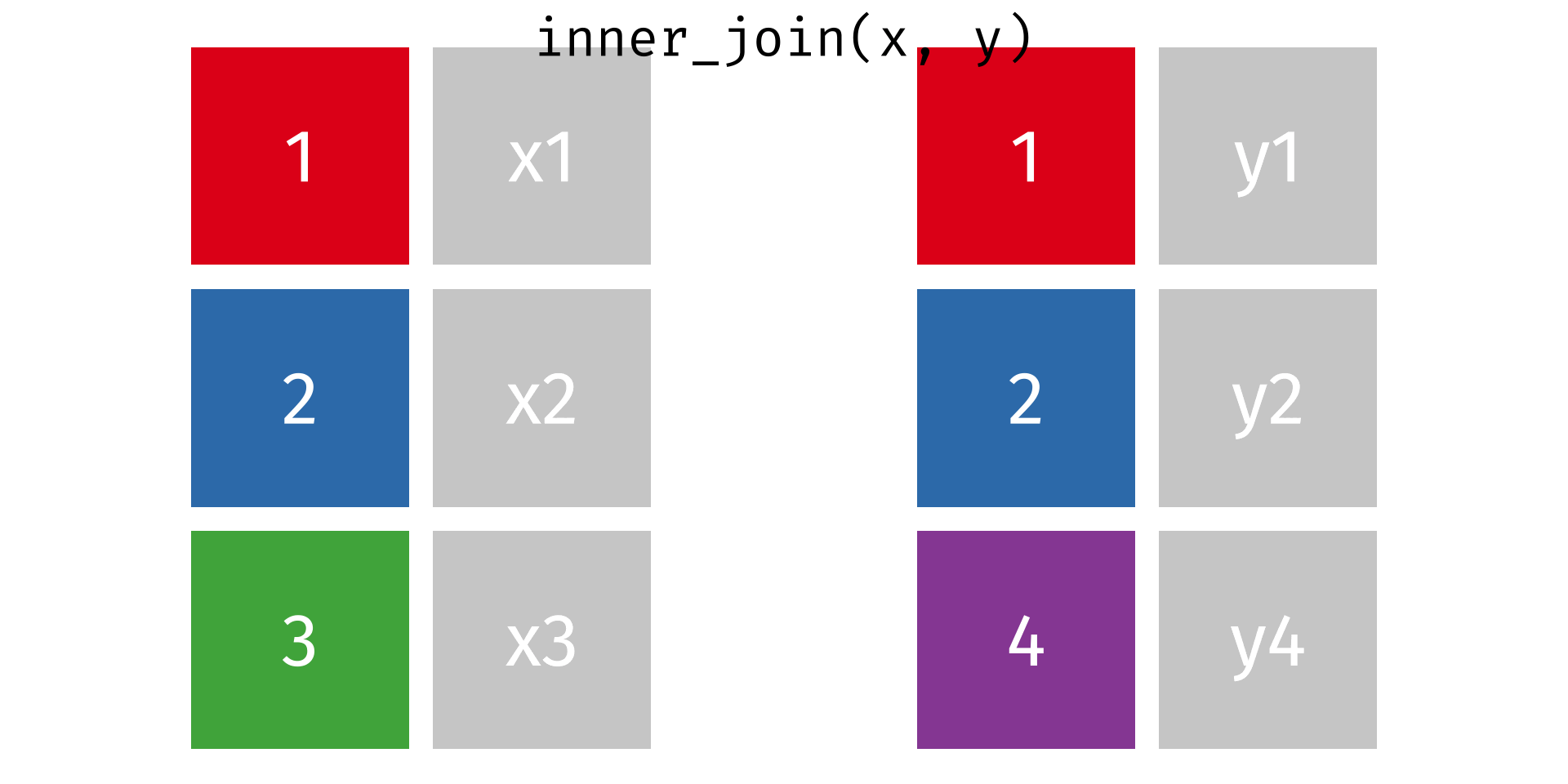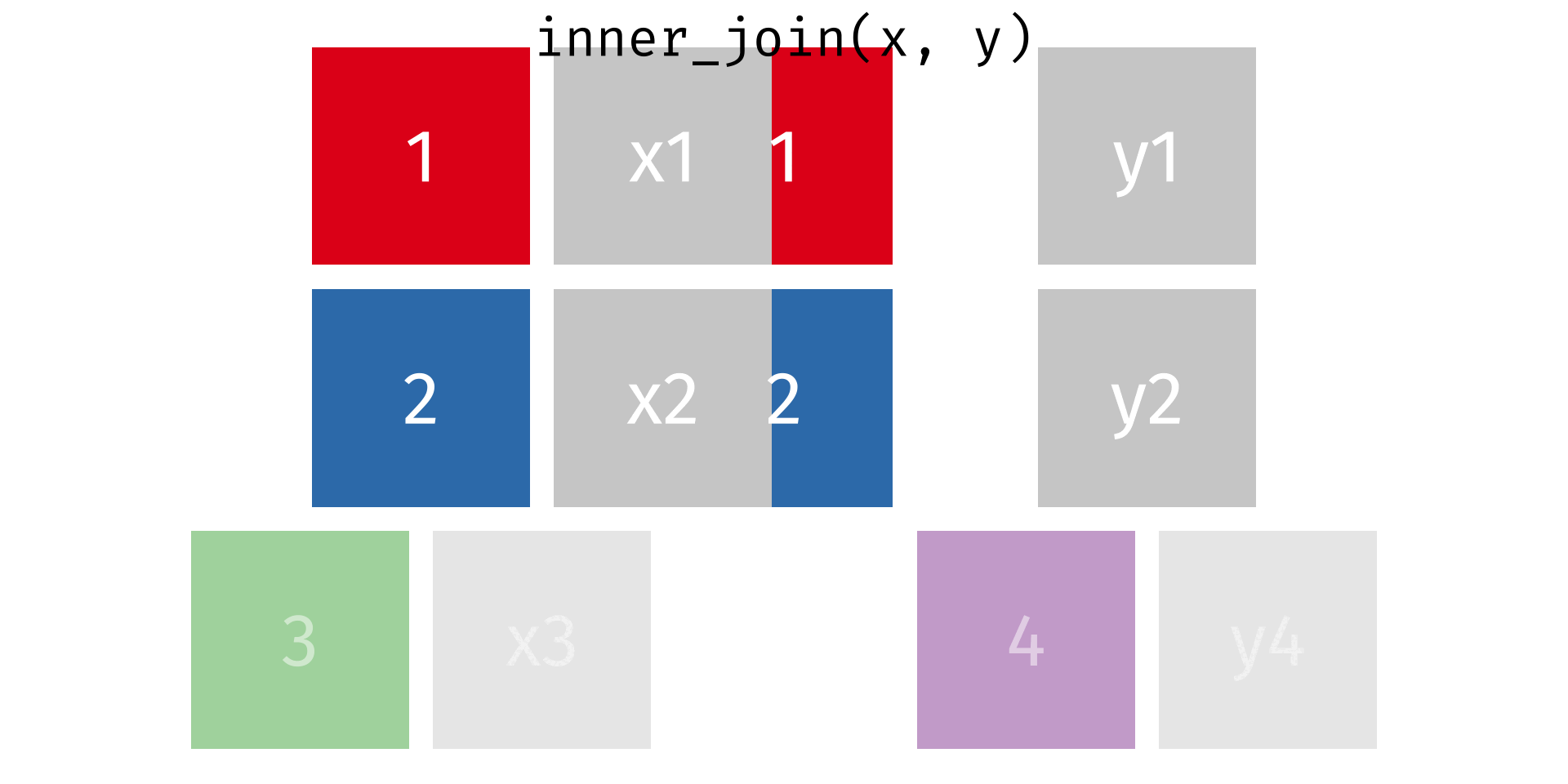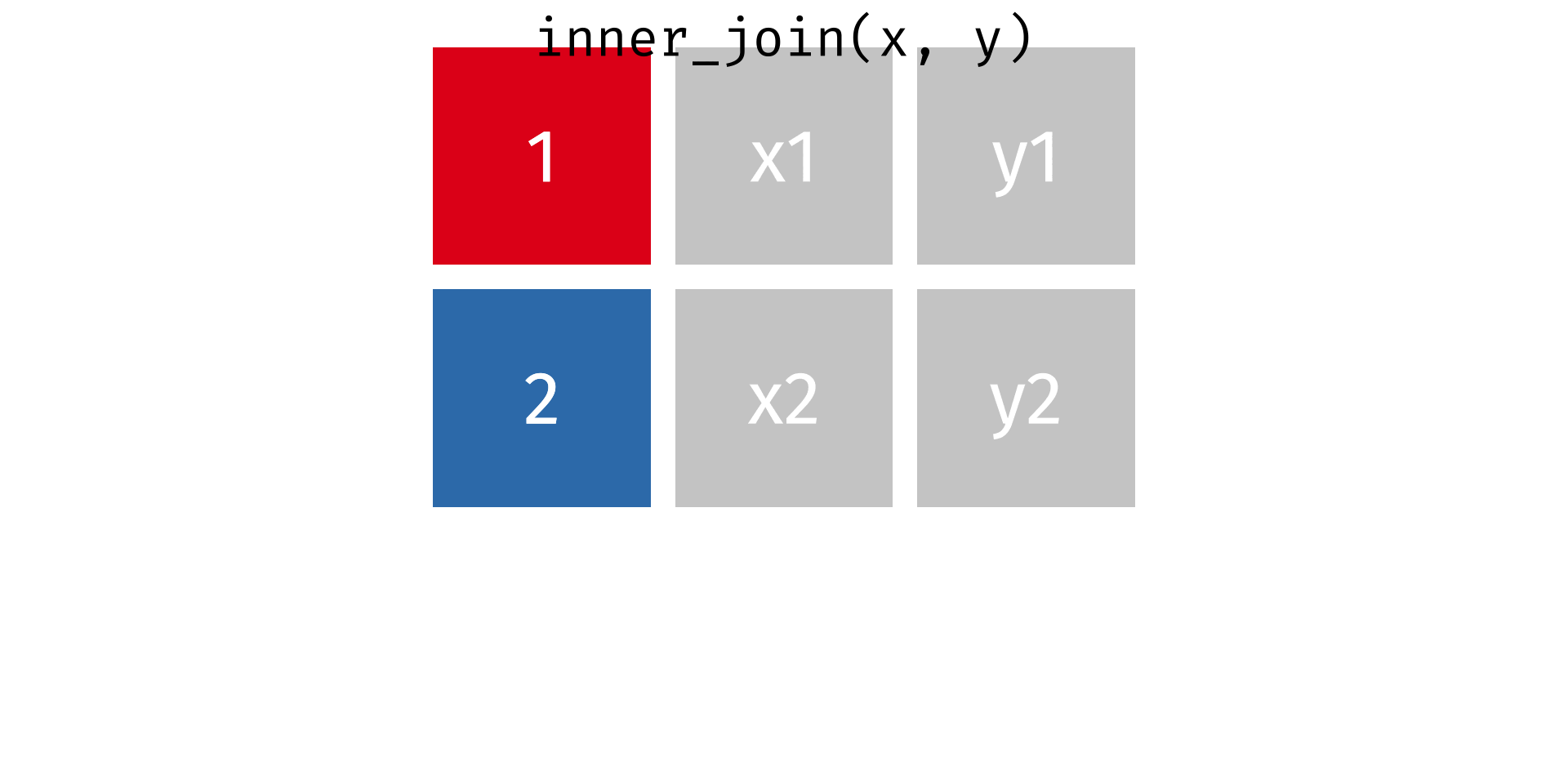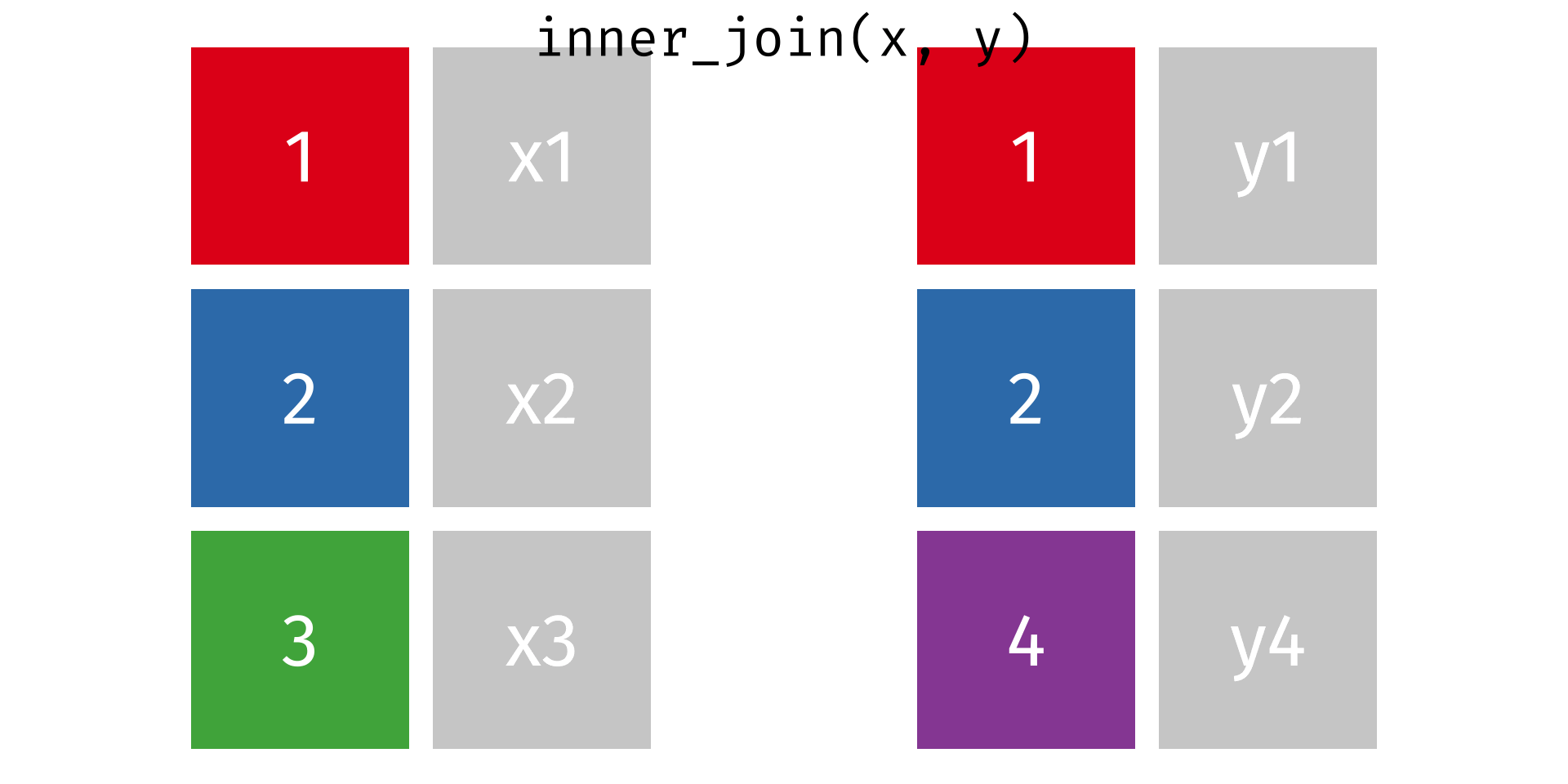
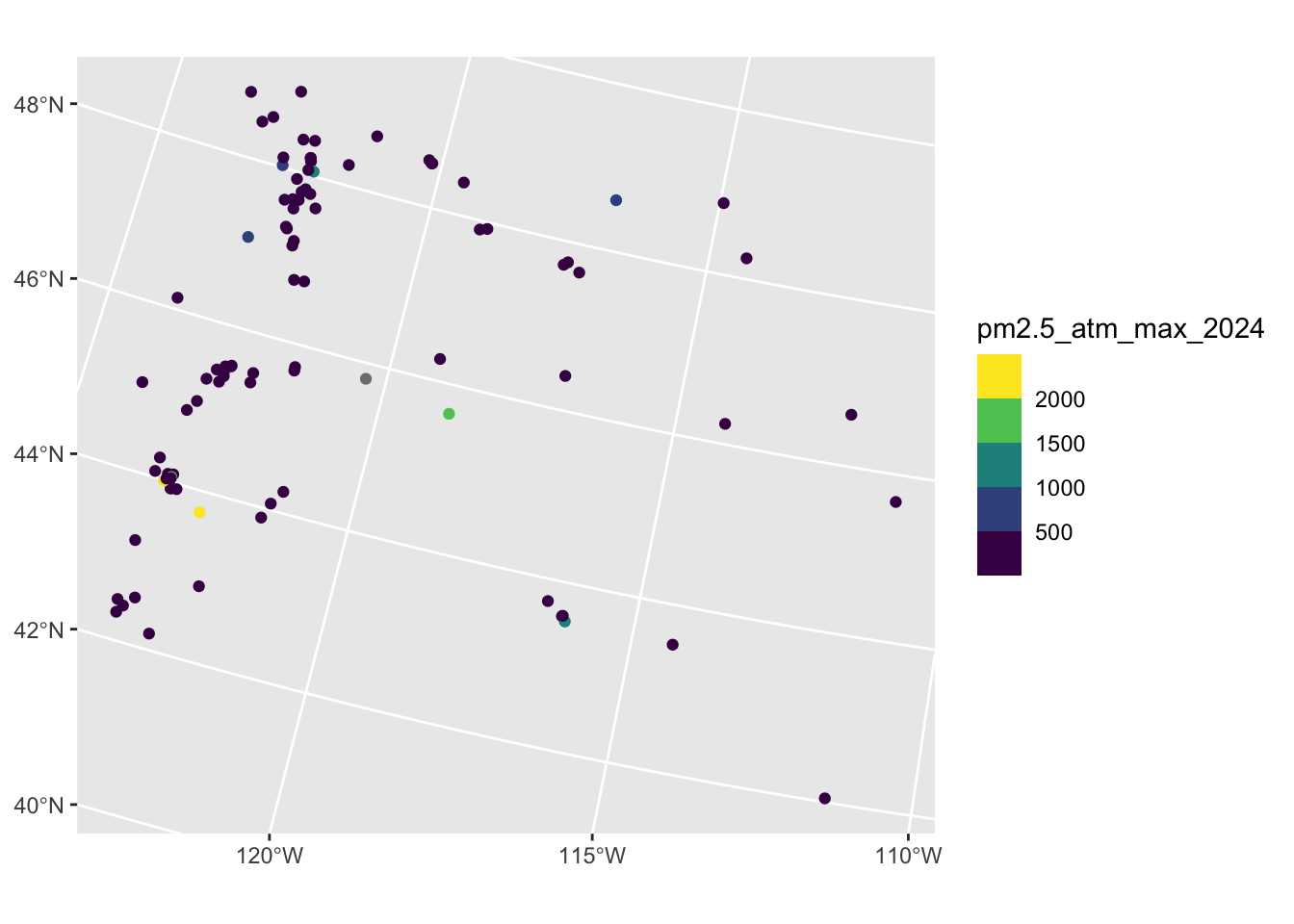
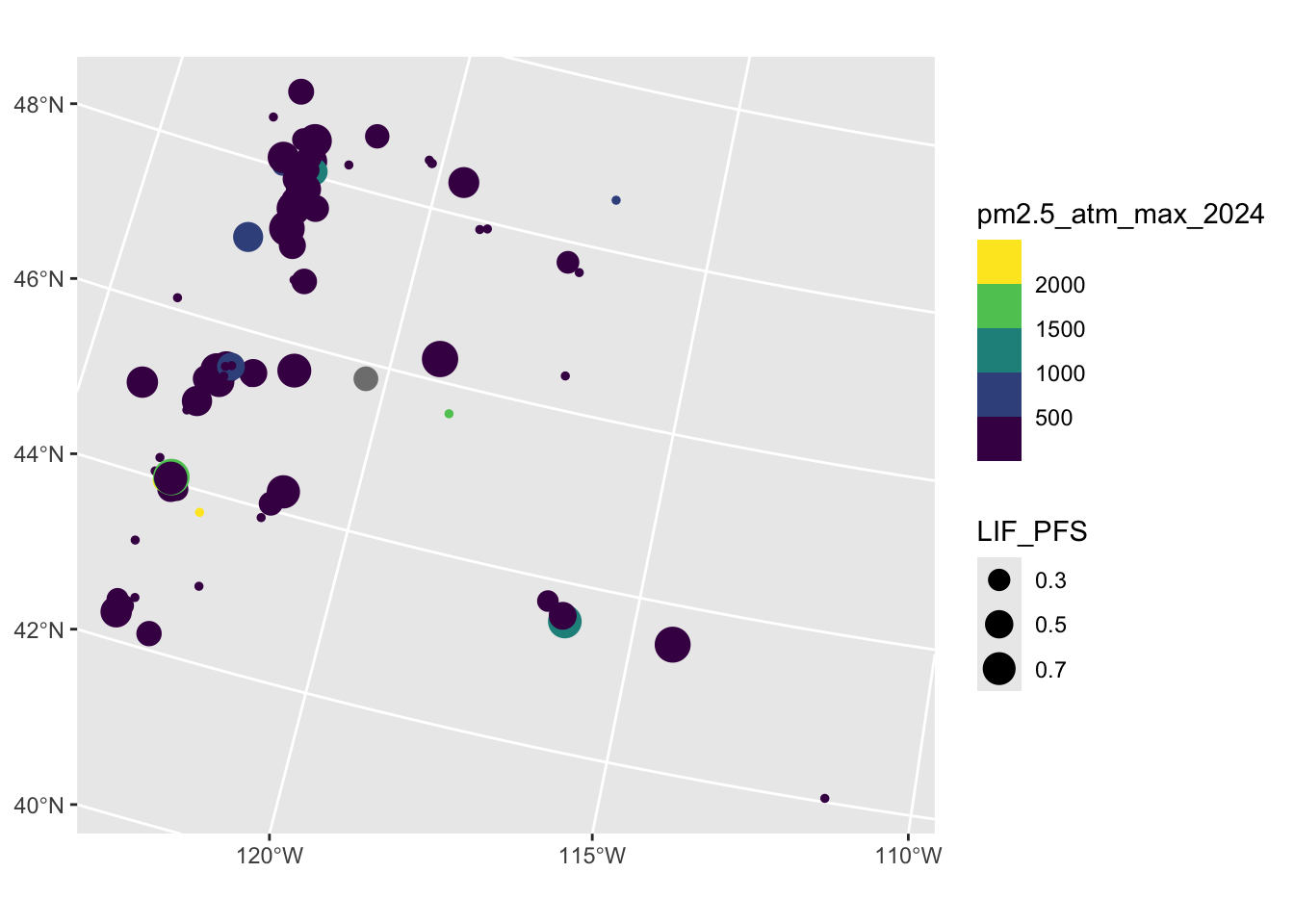
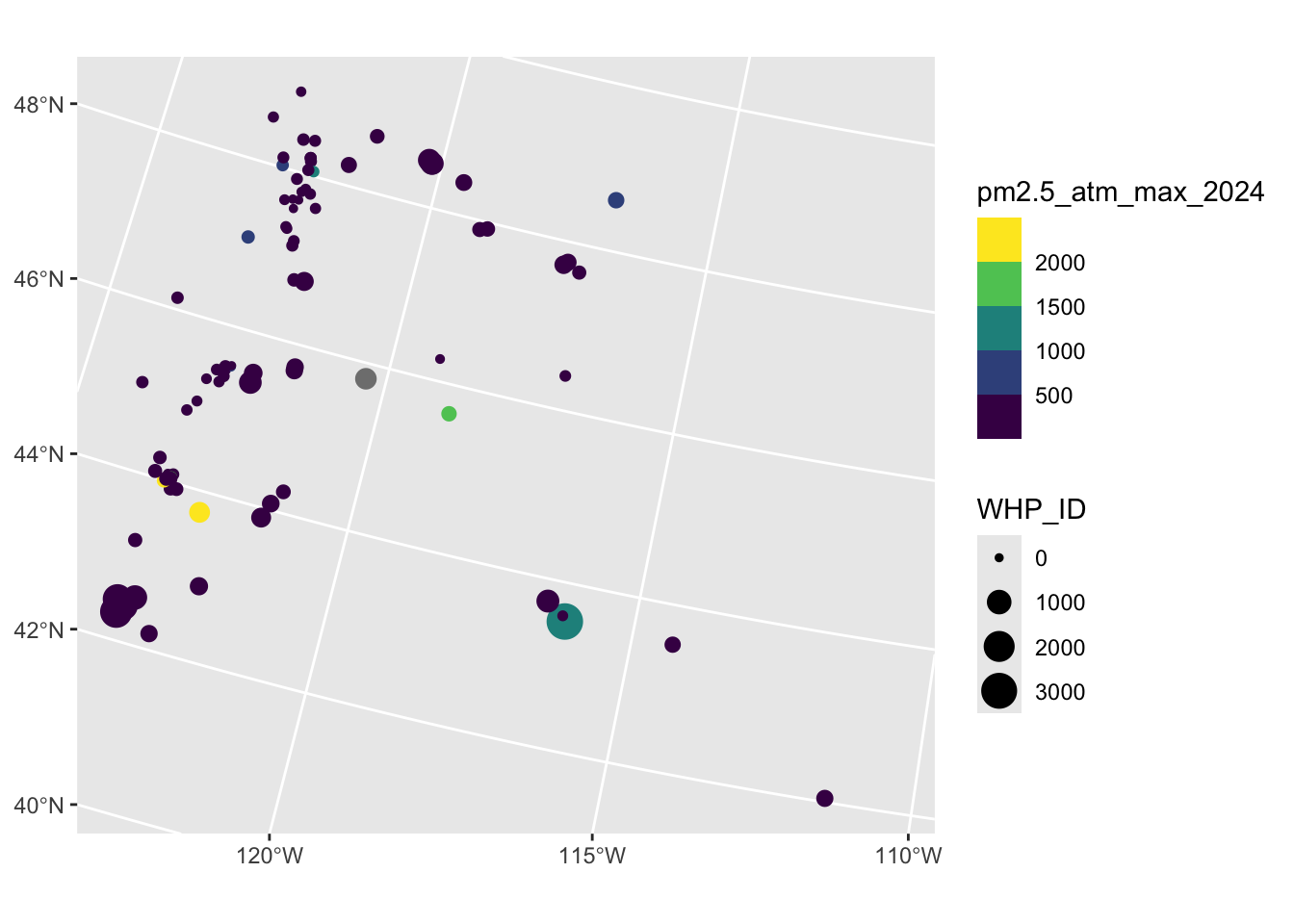
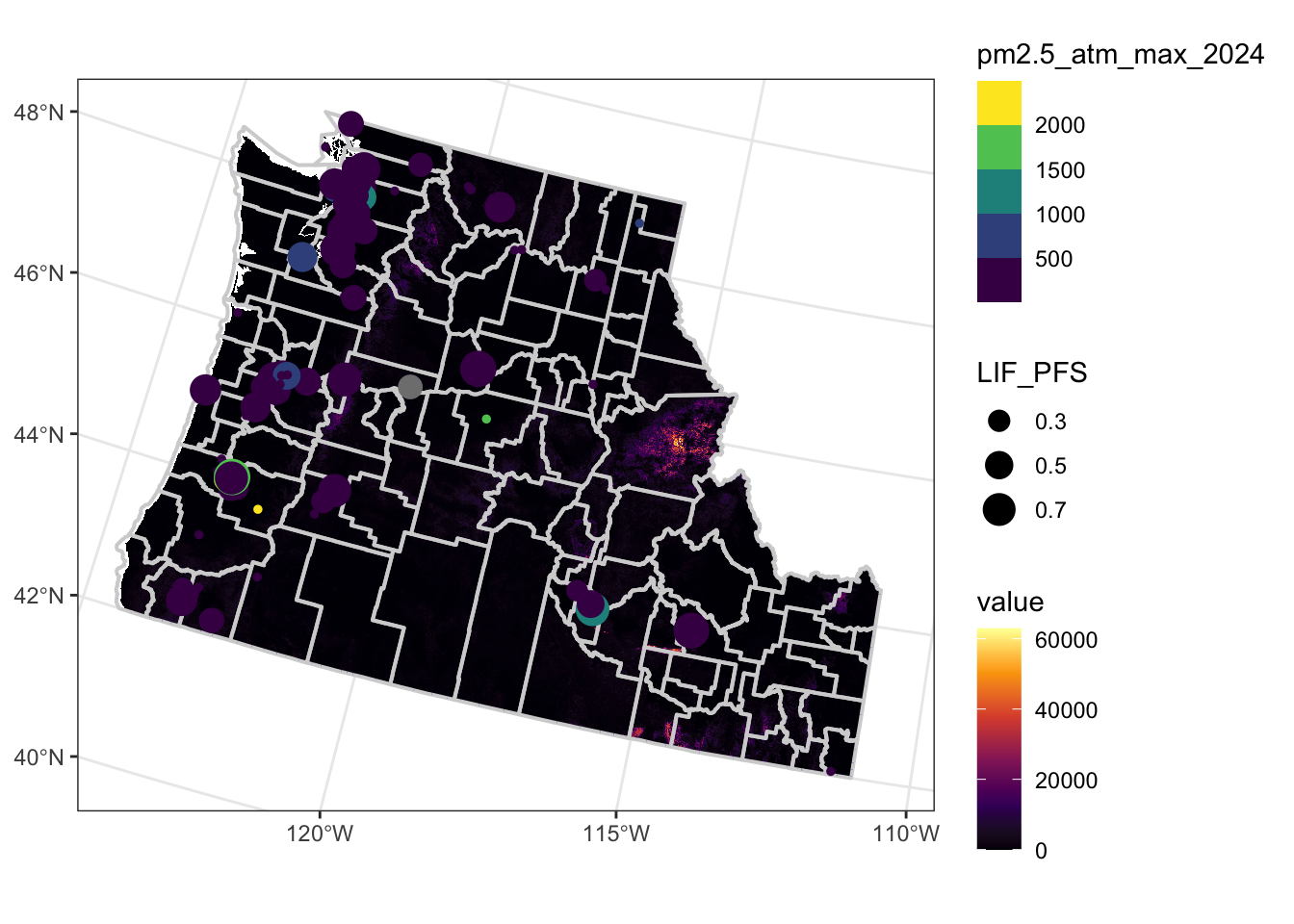
Rows: 2,590
Columns: 124
$ GEOID10 <chr> "16019970700", "16025970100", "16027021700", "16027020700",…
$ SF <chr> "Idaho", "Idaho", "Idaho", "Idaho", "Idaho", "Idaho", "Idah…
$ CF <chr> "Bonneville County", "Camas County", "Canyon County", "Cany…
$ DF_PFS <dbl> 0.44, 0.67, 0.74, 0.48, 0.75, 0.80, 0.76, 0.72, 0.76, 0.52,…
$ AF_PFS <dbl> 0.73, 0.65, 0.86, 0.63, 0.79, 0.88, 0.86, 0.79, 0.80, 0.65,…
$ HDF_PFS <dbl> 0.57, 0.79, 0.69, 0.47, 0.72, 0.55, 0.53, 0.71, 0.74, 0.45,…
$ DSF_PFS <dbl> 0.53, 0.00, 0.58, 0.46, 0.12, 0.79, 0.68, 0.27, 0.06, 0.11,…
$ EBF_PFS <dbl> 0.63, 0.95, 0.54, 0.30, 0.63, 0.81, 0.81, 0.44, 0.73, 0.52,…
$ EALR_PFS <dbl> 0.55, 0.83, 0.63, 0.63, 0.65, NA, 0.41, 0.64, 0.65, 0.64, 0…
$ EBLR_PFS <dbl> 0.19, 0.93, 0.05, 0.55, 0.09, 0.09, 0.08, 0.09, 0.47, 0.17,…
$ EPLR_PFS <dbl> 0.80, 0.99, 0.09, 0.10, 0.11, 0.31, 0.08, 0.09, 0.10, 0.09,…
$ HBF_PFS <dbl> 0.69, 0.60, 0.53, 0.07, 0.46, 0.86, 0.93, 0.22, 0.53, 0.28,…
$ LLEF_PFS <dbl> 0.73, 0.48, 0.67, 0.29, 0.51, 0.87, 0.62, 0.38, 0.32, 0.26,…
$ LIF_PFS <dbl> 0.54, 0.54, 0.53, 0.12, 0.48, 0.61, 0.88, 0.46, 0.41, 0.32,…
$ LMI_PFS <dbl> 0.81, 0.75, 0.61, 0.25, 0.55, 0.92, 0.95, 0.44, 0.42, 0.35,…
$ PM25F_PFS <dbl> 0.11, 0.00, 0.68, 0.63, 0.42, 0.66, 0.69, 0.57, 0.26, 0.40,…
$ HSEF <dbl> 0.14, 0.06, 0.20, 0.08, 0.21, 0.24, 0.31, 0.18, 0.18, 0.09,…
$ P100_PFS <dbl> 0.83, 0.63, 0.47, 0.24, 0.60, 0.72, 0.93, 0.27, 0.39, 0.33,…
$ P200_I_PFS <dbl> 0.86, 0.71, 0.81, 0.39, 0.75, 0.98, 0.91, 0.49, 0.67, 0.54,…
$ AJDLI_ET <int> 1, 1, 1, 0, 1, 1, 1, 0, 1, 1, 0, 1, 1, 1, 0, 0, 1, 0, 0, 0,…
$ LPF_PFS <dbl> 0.75, 0.52, 0.26, 0.30, 0.65, 0.62, 0.63, 0.40, 0.51, 0.36,…
$ KP_PFS <dbl> 0.86, 0.21, 0.84, 0.21, 0.61, 0.57, 0.21, 0.43, 0.90, 0.21,…
$ NPL_PFS <dbl> 0.09, 0.05, 0.06, 0.08, 0.03, 0.08, 0.05, 0.05, 0.08, 0.10,…
$ RMP_PFS <dbl> 0.49, 0.01, 0.56, 0.84, 0.26, 0.96, 0.58, 0.36, 0.10, 0.20,…
$ TSDF_PFS <dbl> 0.22, 0.01, 0.14, 0.42, 0.07, 0.55, 0.13, 0.11, 0.10, 0.19,…
$ TPF <dbl> 5589, 1048, 11701, 3901, 5059, 4681, 3006, 7423, 6653, 4693…
$ TF_PFS <dbl> 0.55, 0.06, 0.54, 0.53, 0.28, 0.71, 0.83, 0.24, 0.05, 0.09,…
$ UF_PFS <dbl> 0.61, 0.16, 0.64, 0.33, 0.66, 0.86, 0.80, 0.45, 0.47, 0.61,…
$ WF_PFS <dbl> 0.03, 0.11, 0.98, 0.24, 0.79, 0.98, 0.98, 0.85, 0.42, 0.04,…
$ UST_PFS <dbl> 0.59, 0.02, 0.44, 0.31, 0.17, 0.73, 0.83, 0.13, 0.06, 0.18,…
$ N_WTR <int> 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ N_WKFC <int> 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ N_CLT <int> 0, 1, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0,…
$ N_ENY <int> 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0,…
$ N_TRN <int> 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0,…
$ N_HSG <int> 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0,…
$ N_PLN <int> 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ N_HLTH <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ SN_C <int> 0, 1, 1, 0, 0, 1, 1, 0, 1, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0,…
$ SN_T <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ DLI <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ ALI <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ PLHSE <int> 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ LMILHSE <int> 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ ULHSE <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ EPL_ET <int> 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ EAL_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0,…
$ EBL_ET <int> 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ EB_ET <int> 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0,…
$ PM25_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ DS_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ TP_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ LPP_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ HRS_ET <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ KP_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0,…
$ HB_ET <int> 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ RMP_ET <int> 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ NPL_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ TSDF_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ WD_ET <int> 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ UST_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ DB_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ A_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ HD_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ LLE_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ UN_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ LISO_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ POV_ET <int> 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ LMI_ET <int> 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ IA_LMI_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ IA_UN_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ IA_POV_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ TC <dbl> 0, 3, 1, 0, 0, 4, 5, 0, 2, 0, 0, 3, 0, 0, 0, 0, 1, 0, 0, 0,…
$ CC <dbl> 0, 2, 1, 0, 0, 4, 4, 0, 2, 0, 0, 3, 0, 0, 0, 0, 1, 0, 0, 0,…
$ IAULHSE <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ IAPLHSE <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ IALMILHSE <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ IALMIL_76 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ IAPLHS_77 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ IAULHS_78 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ LHE <int> 1, 0, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0,…
$ IALHE <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ IAHSEF <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ N_CLT_EOMI <int> 0, 1, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0,…
$ N_ENY_EOMI <int> 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0,…
$ N_TRN_EOMI <int> 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0,…
$ N_HSG_EOMI <int> 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0,…
$ N_PLN_EOMI <int> 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ N_WTR_EOMI <int> 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ N_HLTH_88 <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ N_WKFC_89 <int> 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ FPL200S <int> 1, 1, 1, 0, 1, 1, 1, 0, 1, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0,…
$ N_WKFC_91 <int> 1, 0, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0,…
$ TD_ET <int> 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0,…
$ TD_PFS <dbl> 0.67, 0.78, 0.60, 0.63, 0.85, 0.35, 0.29, 0.94, 0.91, 0.78,…
$ FLD_PFS <dbl> 0.83, 0.88, 0.43, 0.24, 0.82, 0.93, 0.97, 0.44, 0.49, 0.71,…
$ WFR_PFS <dbl> 0.70, 0.82, 0.33, 0.87, 0.80, 0.33, 0.83, 0.77, 0.81, 0.78,…
$ FLD_ET <int> 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ WFR_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ ADJ_ET <int> 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ IS_PFS <dbl> 0.64, 0.16, 0.41, 0.53, 0.86, 0.46, 0.51, 0.70, 0.82, 0.86,…
$ IS_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 0, 1,…
$ AML_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ FUDS_RAW <chr> NA, NA, NA, NA, NA, NA, NA, NA, "0", NA, NA, NA, NA, NA, NA…
$ FUDS_ET <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ IMP_FLG <chr> "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0",…
$ DM_B <dbl> 0.04, 0.00, 0.00, 0.00, 0.00, 0.00, 0.00, 0.00, 0.00, 0.01,…
$ DM_AI <dbl> 0.01, 0.00, 0.01, 0.00, 0.00, 0.02, 0.01, 0.00, 0.00, 0.00,…
$ DM_A <dbl> 0.00, 0.00, 0.00, 0.01, 0.00, 0.00, 0.00, 0.00, 0.00, 0.00,…
$ DM_HI <dbl> 0.00, 0.00, 0.01, 0.00, 0.00, 0.00, 0.00, 0.00, 0.00, 0.00,…
$ DM_T <dbl> 0.06, 0.00, 0.06, 0.04, 0.04, 0.06, 0.03, 0.01, 0.02, 0.00,…
$ DM_W <dbl> 0.62, 0.98, 0.59, 0.88, 0.71, 0.54, 0.40, 0.81, 0.83, 0.85,…
$ DM_H <dbl> 0.27, 0.01, 0.36, 0.07, 0.26, 0.42, 0.54, 0.16, 0.15, 0.11,…
$ DM_O <dbl> 0.11, 0.00, 0.15, 0.02, 0.11, 0.12, 0.27, 0.03, 0.02, 0.02,…
$ AGE_10 <dbl> 0.18, 0.16, 0.15, 0.12, 0.12, 0.15, 0.17, 0.10, 0.10, 0.14,…
$ AGE_MIDDLE <dbl> 0.71, 0.65, 0.71, 0.71, 0.68, 0.77, 0.72, 0.71, 0.76, 0.66,…
$ AGE_OLD <dbl> 0.10, 0.18, 0.13, 0.15, 0.18, 0.07, 0.09, 0.18, 0.12, 0.18,…
$ TA_COU_116 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ TA_COUNT_C <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ TA_PERC <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ TA_PERC_FE <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ UI_EXP <chr> "Nation", "Nation", "Nation", "Nation", "Nation", "Nation",…
$ THRHLD <dbl> 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21, 21,…
$ geometry <MULTIPOLYGON [m]> MULTIPOLYGON (((-1283132 23..., MULTIPOLYGON (…
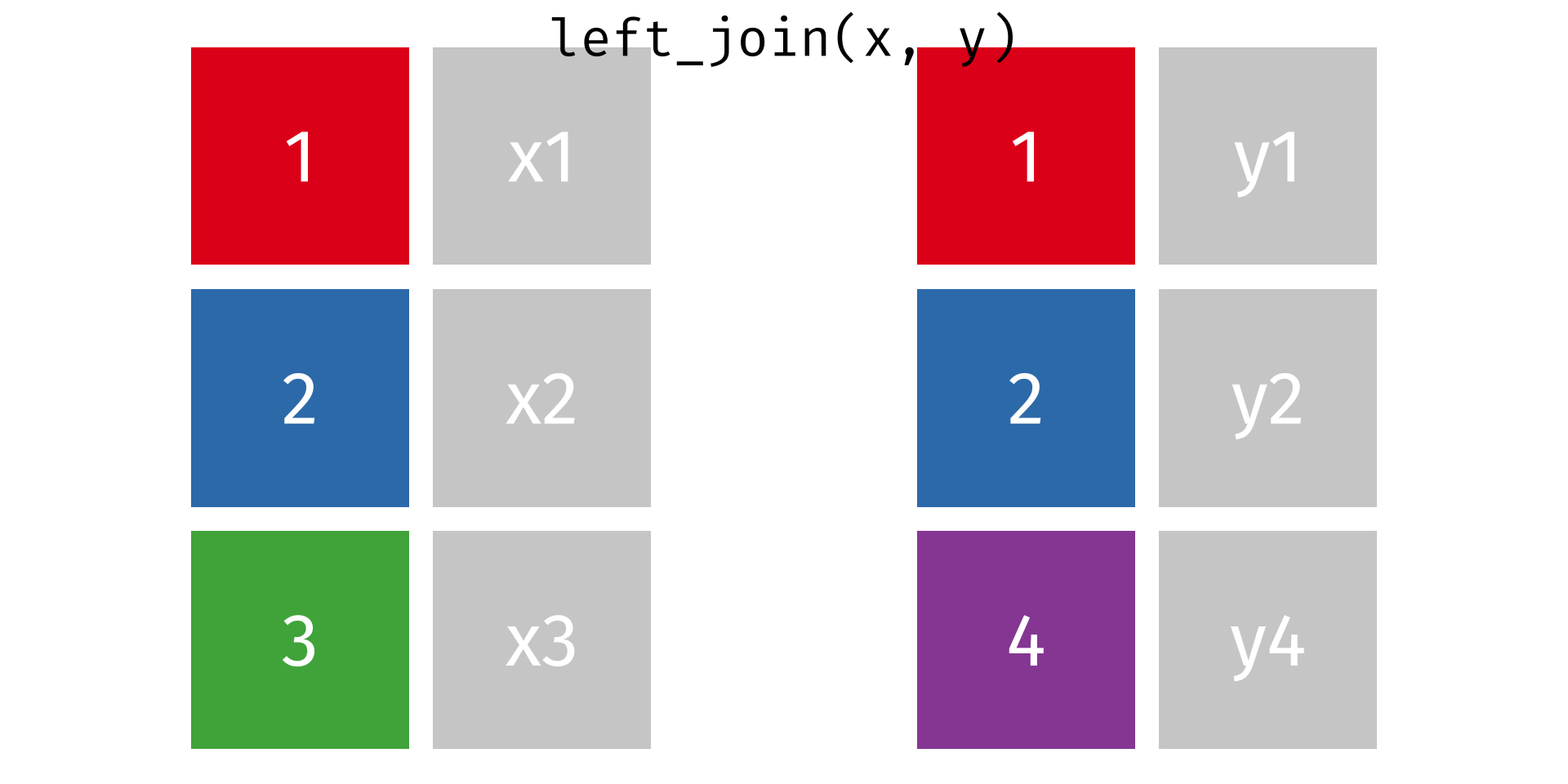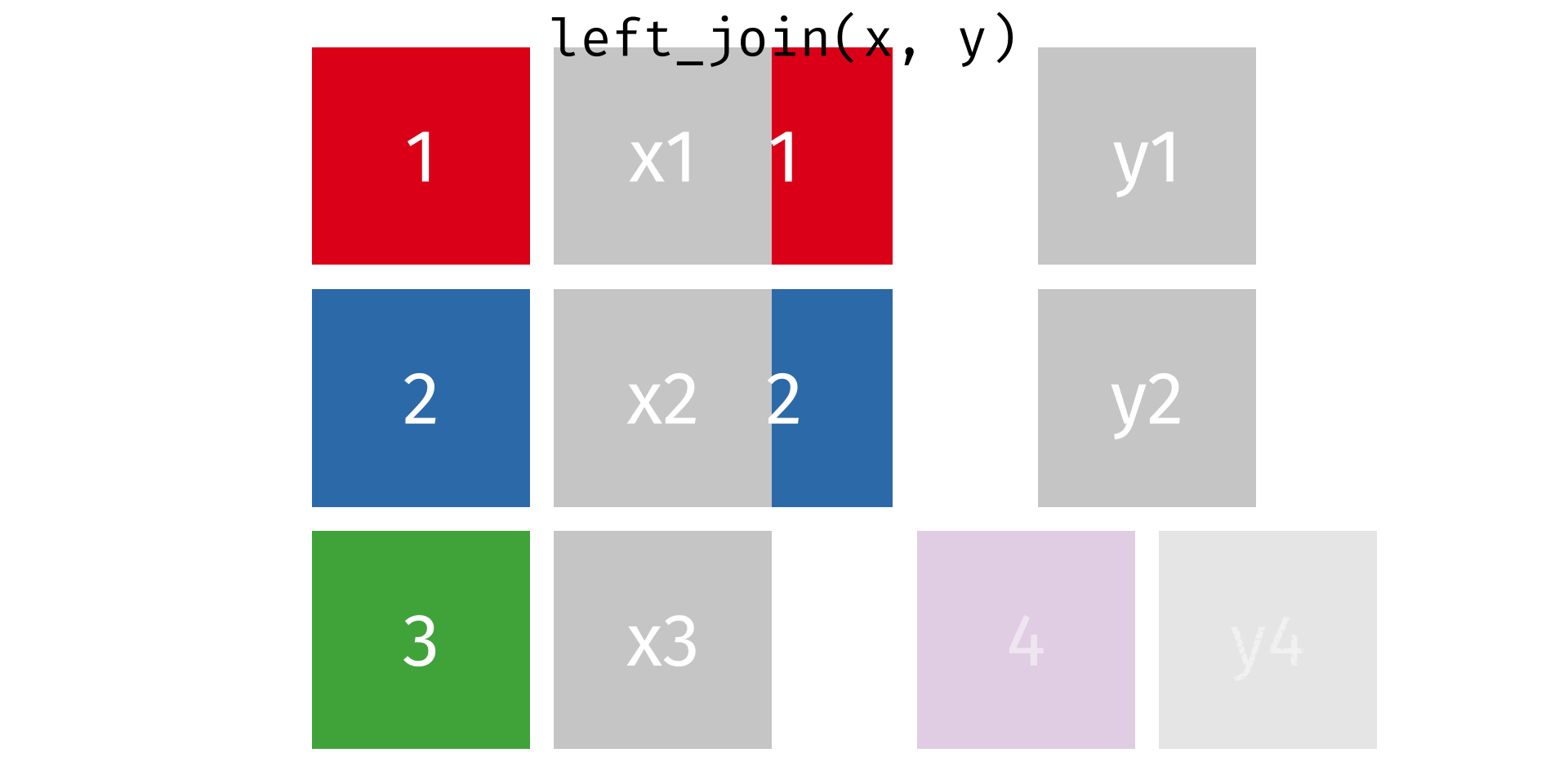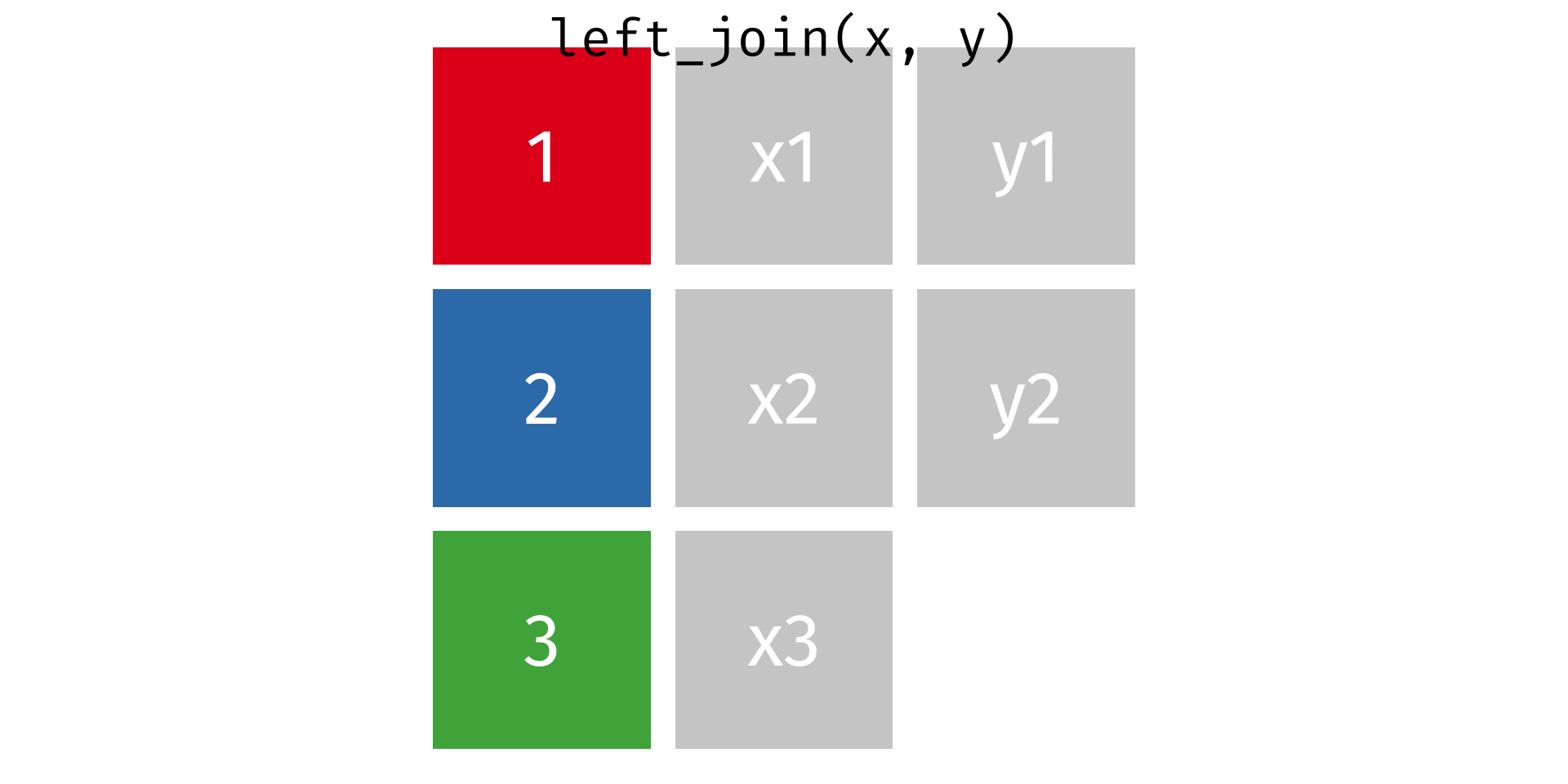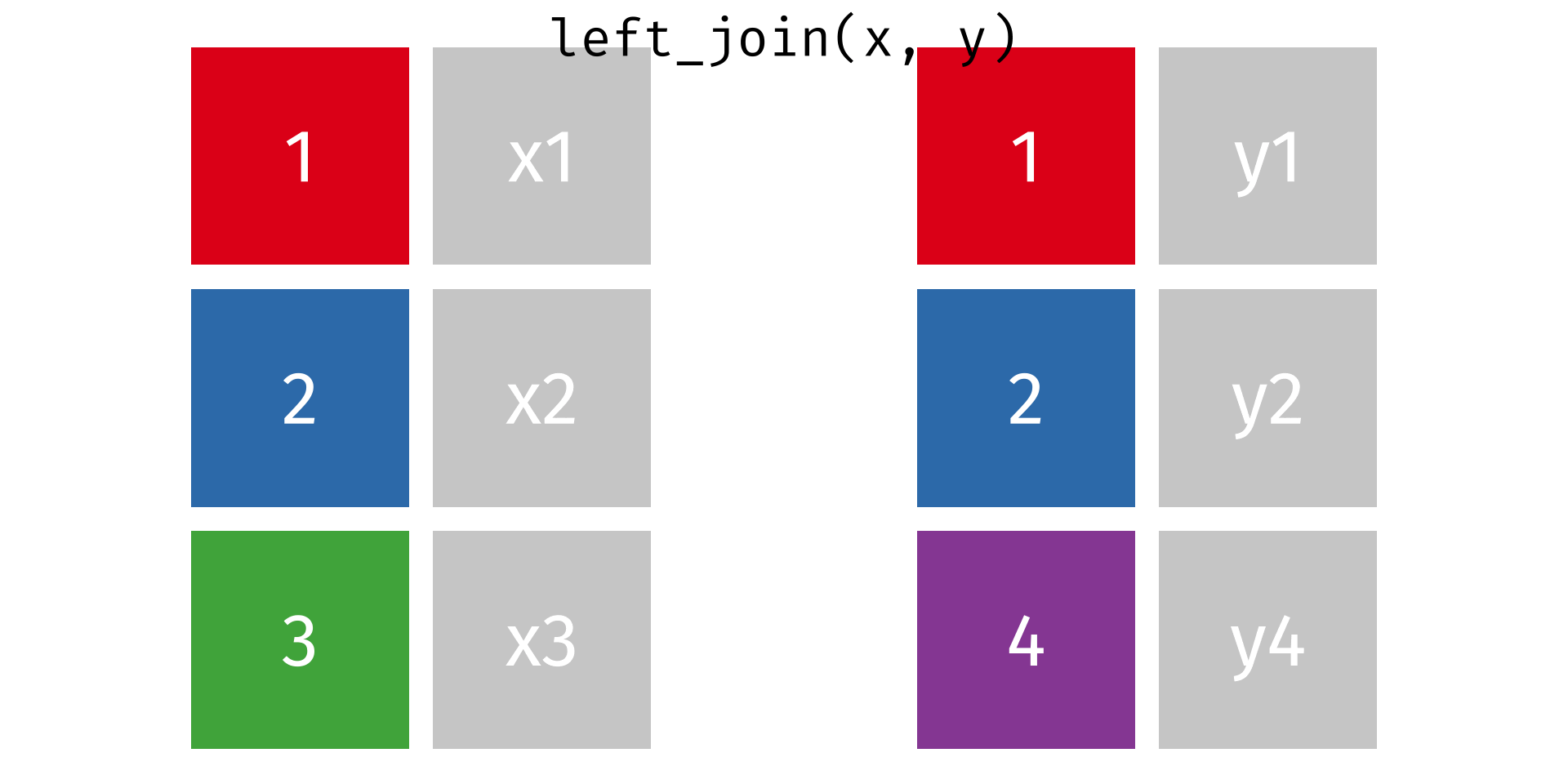 One thing you’ll notice about our tabular dataset (
One thing you’ll notice about our tabular dataset (