Rows: 435
Columns: 9
$ id <dbl> 7494, 7494, 7494, 7494, 7494, 7494, 7494, 7494, 7494, 7494…
$ time_stamp <dttm> 2025-04-01, 2024-04-01, 2021-03-01, 2022-11-01, 2022-07-0…
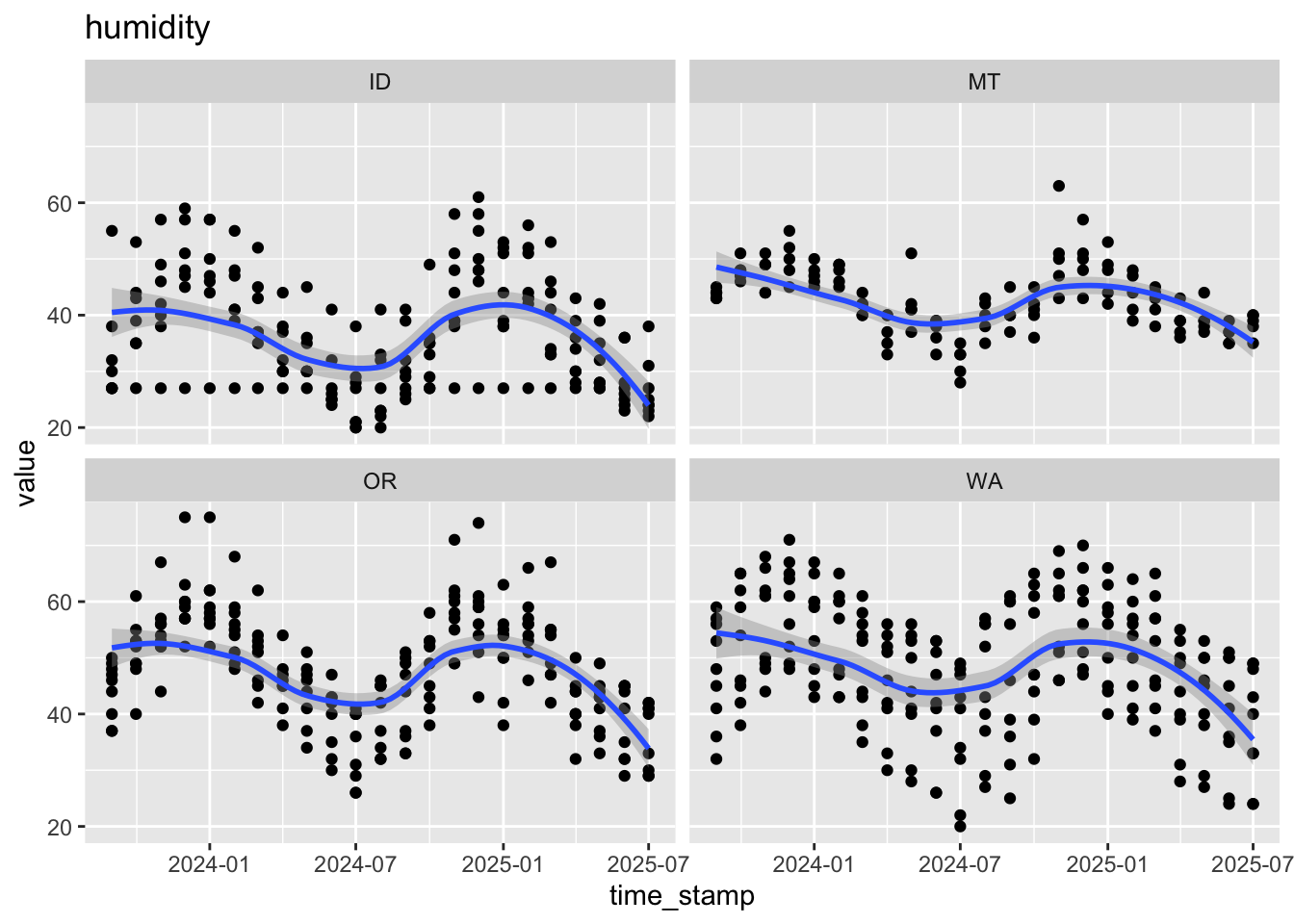
$ humidity <dbl> 30, 30, 30, 41, 20, 27, 21, 30, 35, 29, 32, 24, 33, 46, 30…
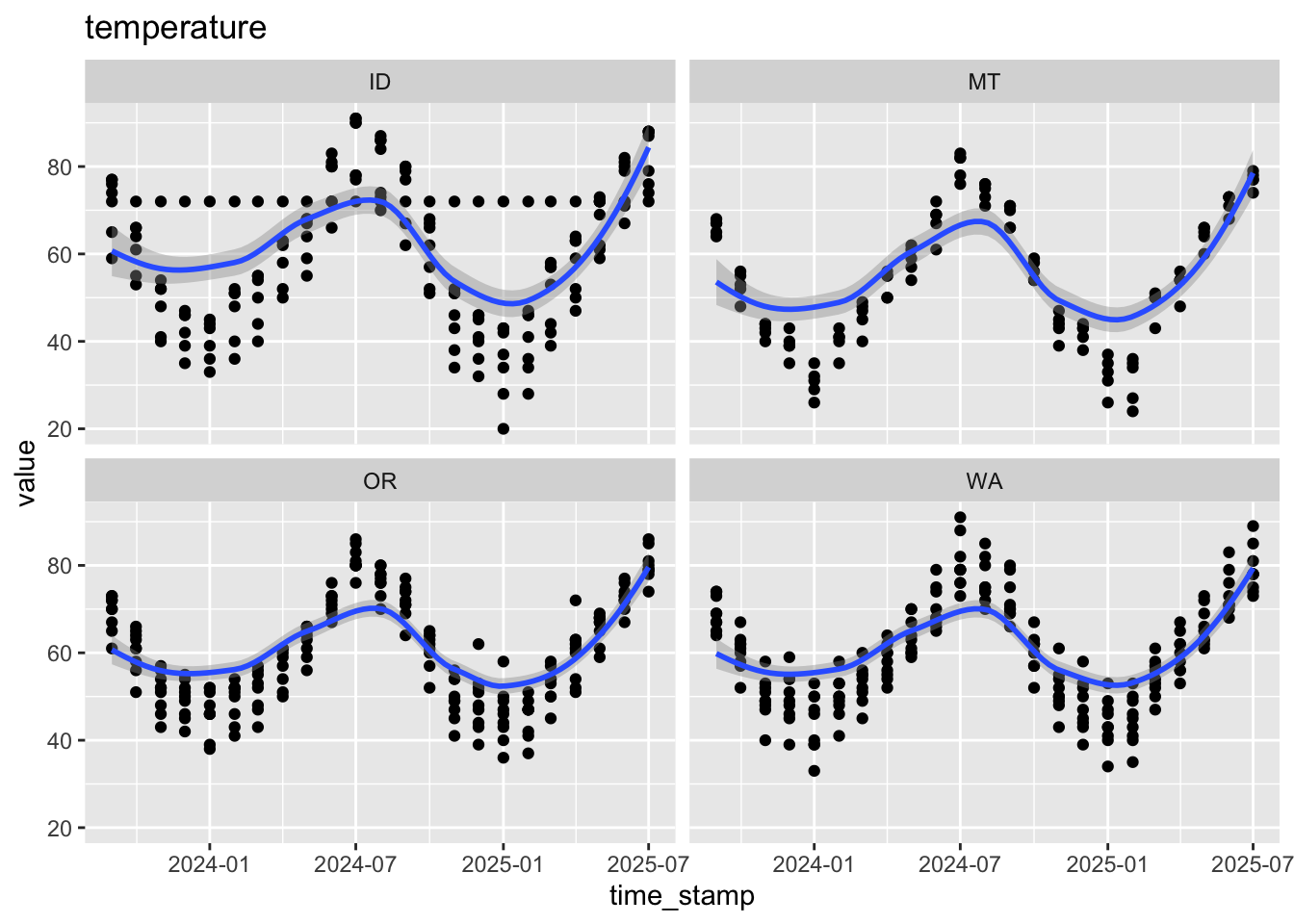
$ temperature <dbl> 63, 63, 56, 45, 91, 83, 91, 76, 66, 79, 57, 82, 66, 45, 86…
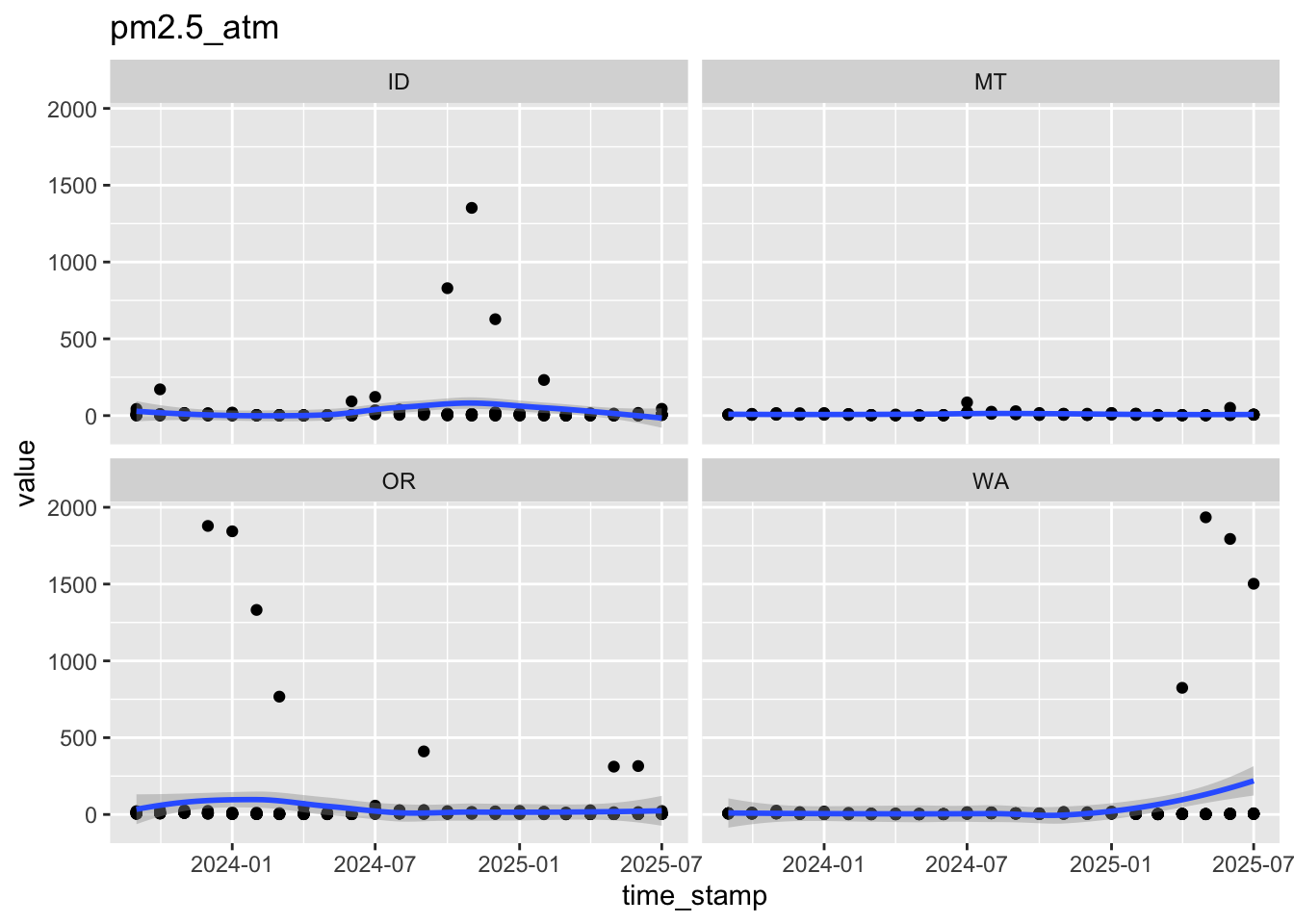
$ pm2.5_atm <dbl> 3.7, 2.2, 4.7, 18.8, 7.3, 27.4, 30.0, 23.1, 1.8, 20.1, 3.9…
$ pm2.5_cf_1 <dbl> 3.7, 2.2, 4.8, 20.2, 7.5, 33.8, 38.3, 31.3, 1.8, 25.6, 4.0…
$ name <chr> "MesaVista", "MesaVista", "MesaVista", "MesaVista", "MesaV…
$ date_up <dttm> 2018-02-10 03:05:11, 2018-02-10 03:05:11, 2018-02-10 03:0…
$ uptime <dbl> 42661, 42661, 42661, 42661, 42661, 42661, 42661, 42661, 42…
Rows: 327
Columns: 9
$ id <dbl> 36673, 36673, 36673, 36673, 36673, 36673, 36673, 36673, 36…
$ time_stamp <dttm> 2022-08-01, 2020-05-01, 2024-01-01, 2021-08-01, 2021-03-0…
$ humidity <dbl> 37, 45, 47, 46, 40, 48, 49, 41, 47, 48, 39, 40, 47, 44, 50…
$ temperature <dbl> 77, 60, 32, 71, 46, 39, 42, 45, 40, 35, 64, 52, 55, 59, 38…
$ pm2.5_atm <dbl> 12.1, 2.8, 16.4, 29.5, 98.9, 9.4, 10.7, 6.4, 7.3, 8.3, 4.2…
$ pm2.5_cf_1 <dbl> 12.8, 3.0, 17.2, 37.1, 145.6, 9.9, 11.5, 6.7, 7.7, 8.9, 4.…
$ name <chr> "419 S. 4th St", "419 S. 4th St", "419 S. 4th St", "419 S.…
$ date_up <dttm> 2019-08-02 19:28:01, 2019-08-02 19:28:01, 2019-08-02 19:2…
$ uptime <dbl> 15585, 15585, 15585, 15585, 15585, 15585, 15585, 15585, 15…
Rows: 563
Columns: 9
$ id <dbl> 9810, 9810, 9810, 9810, 9810, 9810, 9810, 9810, 9810, 9810…
$ time_stamp <dttm> 2020-11-01, 2021-05-01, 2020-05-01, 2023-12-01, 2020-09-0…
$ humidity <dbl> 45, 32, 38, 52, 30, 43, 22, 49, 35, 38, 38, 51, 25, 46, 37…
$ temperature <dbl> 46, 63, 62, 46, 72, 47, 89, 45, 69, 54, 55, 44, 55, 41, 48…
$ pm2.5_atm <dbl> 23.1, 3.9, 4.5, 23.6, 56.1, 12.6, 31.9, 13.0, 2.7, 6.2, 25…
$ pm2.5_cf_1 <dbl> 27.4, 4.3, 4.6, 28.3, 79.9, 13.7, 40.5, 14.2, 2.7, 6.3, 30…
$ name <chr> "PSU STAR LAB KFP", "PSU STAR LAB KFP", "PSU STAR LAB KFP"…
$ date_up <dttm> 2018-04-11 23:19:19, 2018-04-11 23:19:19, 2018-04-11 23:1…
$ uptime <dbl> 22520, 22520, 22520, 22520, 22520, 22520, 22520, 22520, 22…
Rows: 603
Columns: 9
$ id <dbl> 18161, 18161, 18161, 18161, 18161, 18161, 18161, 18161, 18…
$ time_stamp <dttm> 2020-08-01, 2023-01-01, 2022-03-01, 2024-01-01, 2023-10-0…
$ humidity <dbl> 47, 57, 58, 59, 59, 50, 61, 61, 54, 48, 42, 61, 60, 50, 51…
$ temperature <dbl> 75, 52, 55, 50, 62, 52, 56, 54, 54, 72, 79, 47, 52, 72, 55…
$ pm2.5_atm <dbl> 5.7, 2.8, 4.2, 3.5, 6.4, 3.2, 2.7, 4.6, 3.1, 16.0, 7.5, 4.…
$ pm2.5_cf_1 <dbl> 5.7, 2.8, 4.4, 3.6, 6.4, 3.3, 2.8, 4.7, 3.2, 18.3, 8.0, 4.…
$ name <chr> "Issaquah Highlands", "Issaquah Highlands", "Issaquah High…
$ date_up <dttm> 2018-10-29 22:26:57, 2018-10-29 22:26:57, 2018-10-29 22:2…
$ uptime <dbl> 13394, 13394, 13394, 13394, 13394, 13394, 13394, 13394, 13…